Minimize the entropic Wasserstein distance
minimize_entropic_W.RdMinimize the entropic Wasserstein distance
minimize_entropic_W(
Y,
A = NULL,
AV = NULL,
P_D = NULL,
muStart = NULL,
maxIter = 100L,
minIter = 50L,
sinkhornIter = 100L,
eps = 0.01,
gammaStart = 0.05,
p = 1,
sinkhornTol = 1e-12,
gradDescTol = 1e-12,
fastSinkhorn = TRUE,
pushforwardStart = FALSE,
suppressOutput = FALSE,
WThreshold = 0
)Arguments
- Y
numeric vector of observations
- A
numeric vector of domain values where you expect most of the Y values to lie in
- AV
numeric vector of domain values which you allow the functions in the space you minimize over to take
- P_D
Jacobian of mu -> W_2(mu*D, pi_hat)
- muStart
starting values for minimization
- maxIter
maximum iteration number for gradient descent algorithm
- minIter
minimum iteration number for gradient descent algorithm
- sinkhornIter
maximum Sinkhorn iterations
- eps
entropic regularization constant
- gammaStart
starting gamma value for gradient descent algorithm. Only used in first descent step.
- p
exponent of L_p norm
- sinkhornTol
tolerance for stopping criterion in Sinkhorn algorithm
- gradDescTol
tolerance for stopping criterion in gradient descent algorithm based on euclidian distance to last vector
- fastSinkhorn
logical, controls whether to use the fast Sinkhorn algorithm
- pushforwardStart
if TRUE, starts with nearest neighbour distribution to pi_hat instead of rep(1/n)
- suppressOutput
suppress output messages?
- WThreshold
tolerance for stopping criterion for gradient descent algorithm based on the Wasserstein distance
Examples
library(tidyverse)
#> ── Attaching packages ─────────────────────────────────────── tidyverse 1.3.1 ──
#> ✔ ggplot2 3.3.5 ✔ purrr 0.3.4
#> ✔ tibble 3.1.6 ✔ dplyr 1.0.7
#> ✔ tidyr 1.1.4 ✔ stringr 1.4.0
#> ✔ readr 2.1.1 ✔ forcats 0.5.1
#> ── Conflicts ────────────────────────────────────────── tidyverse_conflicts() ──
#> ✖ dplyr::filter() masks stats::filter()
#> ✖ dplyr::lag() masks stats::lag()
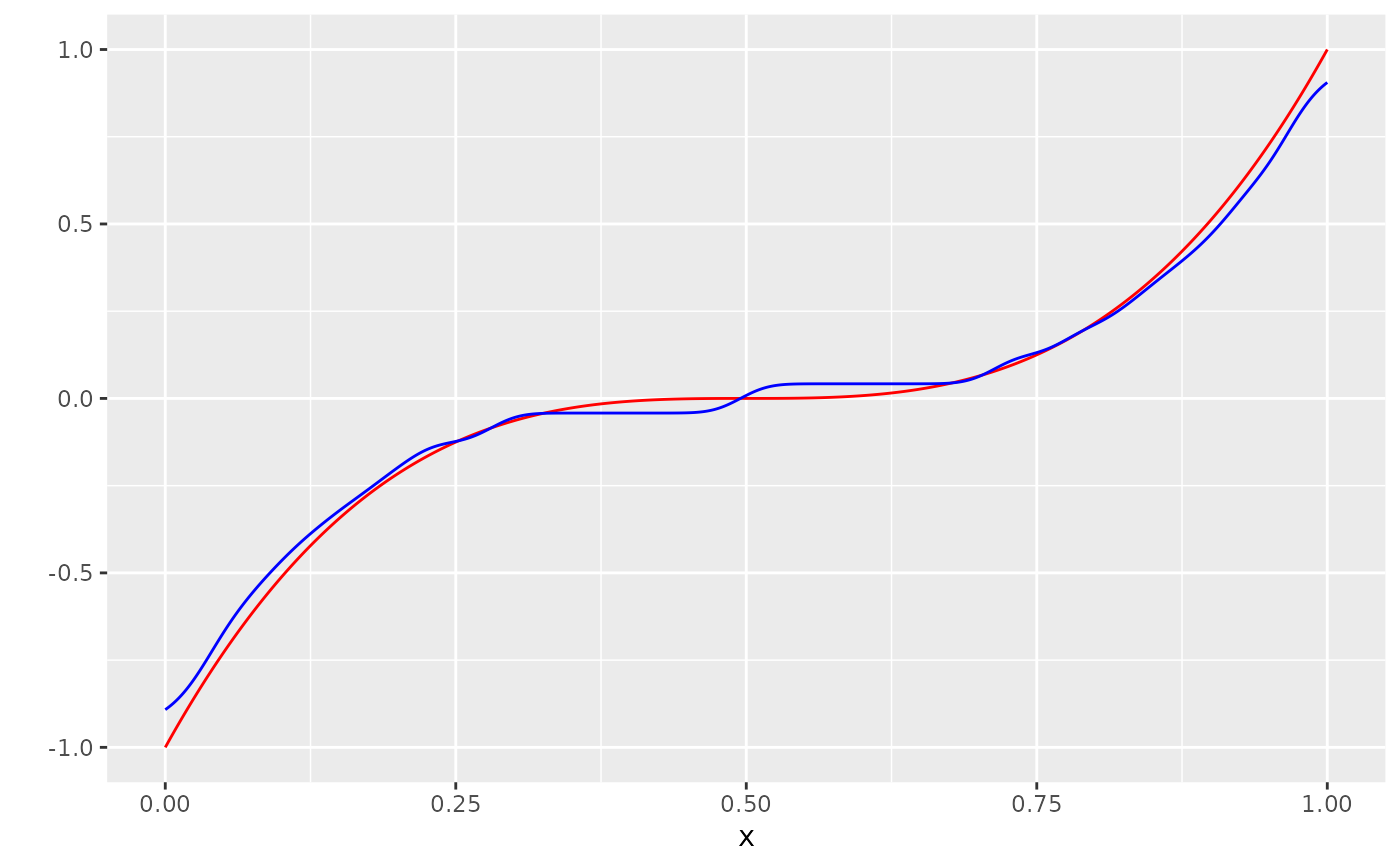
n <- 1000
x <- seq(0, 1, length.out = n)
m <- function(x) (2*(x- 0.5))^3
Y_no_error <- m(x)
varepsilon <- rbernoulli_custom(n, a= -0.3, b= 0.3, p=0.5)
Y <- (Y_no_error + varepsilon) %>% sample(n)
dat <- tibble(x=x, Y=Y, Y_no_error = Y_no_error)
N <- round(sqrt(n))
A <- seq(-1.3, 1.3, length.out = N)
stepsize <- (A[2]-A[1]) / 2
A_V <- A[which(-1 - stepsize <= A & A <= 1 + stepsize)]
p_ber <- function(x) pbernoulli_custom(x, a = -0.3, b = 0.3, p=0.5)
P <- matrix(rep(0, times = (N) * length(A_V)), nrow = N)
P[1, ] <- p_ber(A[2] - stepsize - A_V)
for (i in 2:(N-1)) {
P[i,] <- p_ber(A[i + 1] - stepsize - A_V) - p_ber(A[i] - stepsize - A_V)
}
P[N, ] <- 1 - p_ber(A[N] - stepsize - A_V)
l <-
minimize_entropic_W(Y = Y,
A = A,
AV = A_V,
P_D = P,
suppressOutput = FALSE)
#> Maximum number iterations reached (100).
mu_hat <- list()
mu_hat[["vals"]] <- l$vals
mu_hat[["probs"]] <- l$probs
g_hat <- measure_to_smooth_iso(mu_hat, n)
dat <- bind_cols(dat, g_hat = g_hat)
ggplot(dat, aes(x=x, y=Y_no_error)) +
geom_line(col="red") +
scale_y_continuous("") +
geom_line(aes(y=g_hat), col="blue")